Abstract
Background: Iron deficiency anemia (IDA) is highly prevalent worldwide, especially in premenopausal women during their reproductive years because of iron loss associated with menstruation and pregnancy. In 2019, the global prevalence of anemia was 36.5% among pregnant women, 29.6% in non-pregnant women Although iron is one of the most abundant elements on earth, it is not readily available for colonizing bacteria due to its low solubility in the human body. Hosts and microbiota compete fiercely for iron. Iron deficiency not only causes anemia, but also causes a variety symptoms including dyspnea, general weakness. Also, iron deficiency impact gut microbial health. There are some previous studies on the gut microbiome of patients with IDA. However, the study of the human microbiome is difficult to control for various variables that can influence the microbiome. To the best of our knowledge, there are limited studies regarding know the effects of iron supplementation on the gut microbiota in human.
Aims: The aim of this study is to compare the microbiome of patients with IDA and normal individuals from young women. We also wanted to know the effects of iron supplementation on the gut microbiota.
Methods: We enrolled 31 women in the study, including 15 IDA patients and 16 healthy volunteers. Participants, between 20 and 50 years of age, premenopausal women. All patients in the IDA group were diagnosed with IDA according to WHO diagnostic standards. Control participants were self-reported as healthy and did not suffer from an illness at the time of recruitment. Paticipants didn't use antibiotics and any other medical treatment that influences intestinal microbiota during 3 months before the study start. Stool sampling was conducted according to the Manual of Procedures for Human Microbiome Project Core Microbiome Sampling Protocol by the NIH at the time of study start. Following iron replacement for 3~6 months, the IDA patients performed a blood test to confirm that the anemia had improved, and stool sampling was conducted again at that time. Samples were obtained from 10 IDA patients before and after treatment with iron. 16S rRNA gene-based metagenomic analysis of fecal microbiome was performed.
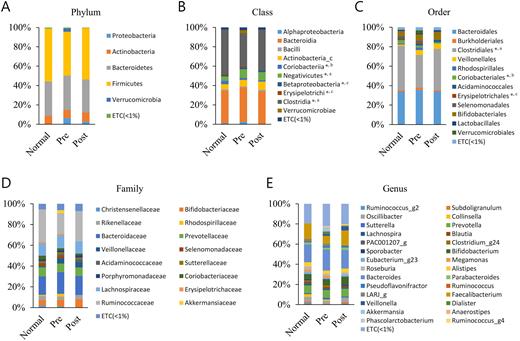
Results: Among the taxa with an average taxonomic composition greater than 1%, five classes (c-Coriobacteria, c-Negativicutes, c-Betaproteobacteria, c-Erysipelotrichi and c-Closteridia) and three orders (o-Clostridiales, o-Coriobacterials and o-Erysipelotrichales) significantly differ in three groups (Figure). Alpha diversity indices showed no differences between the groups and beta set-significance analysis indicated distinctive differences between normal and pre-treatment groups. Moreover, taxonomic biomarkers discovery by linear discriminant analysis Effect Size (LEfSe) analysis between normal and pre-treatment group showed higher effect size of f-Ruminococcaceae, o-Clostridiales, c-Clostridia, g-Faecalibacterium in normal and c-Negativicutes in pre-treatment group. Similarly, LEfSe analysis among treatment groups presented higher effect size of c-Erysipelotrichi, o-Erysipelotrichales, f-Erysipelotrichaceae in pre-treatment group. Whereas, LEfSe analysis between normal and post-treatment groups indicated that c-Coriobacteria, f-Coriobacteriaceae and o-Coriobacteriales were prominent in normal and g-Veillonella and g-Collinsella were significant in Post-treatment group. Finally, functional biomarker discovery by LEfSe analysis showed 15 Kos, 4 modules and two pathways distinctively different among the groups.
Conclusions: In conclusion, there was a significant difference between the gut microbiome of the patients of IDA and that of healthy volunteers. We found a tendency of recovery of gut microbiome after iron supplementation treatment.
Figure. Averaged taxonomic composition in stool of Normal, Pre- and Post-treatment IDA (iron deficiency anemia) groups. Taxonomic relative abundance was classified at the (A) phylum, (B) class, (C) order, (D) family, and (E) genus level, and relative abundances less than 1 % were expressed as ETC. Wilcoxon rank-sum test was used to analyze the significance between the two groups. a: Comparison between Normal and Pre; b: Comparison between Normal and Post; c: Comparison between Pre and Post;*, p<0.05.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal